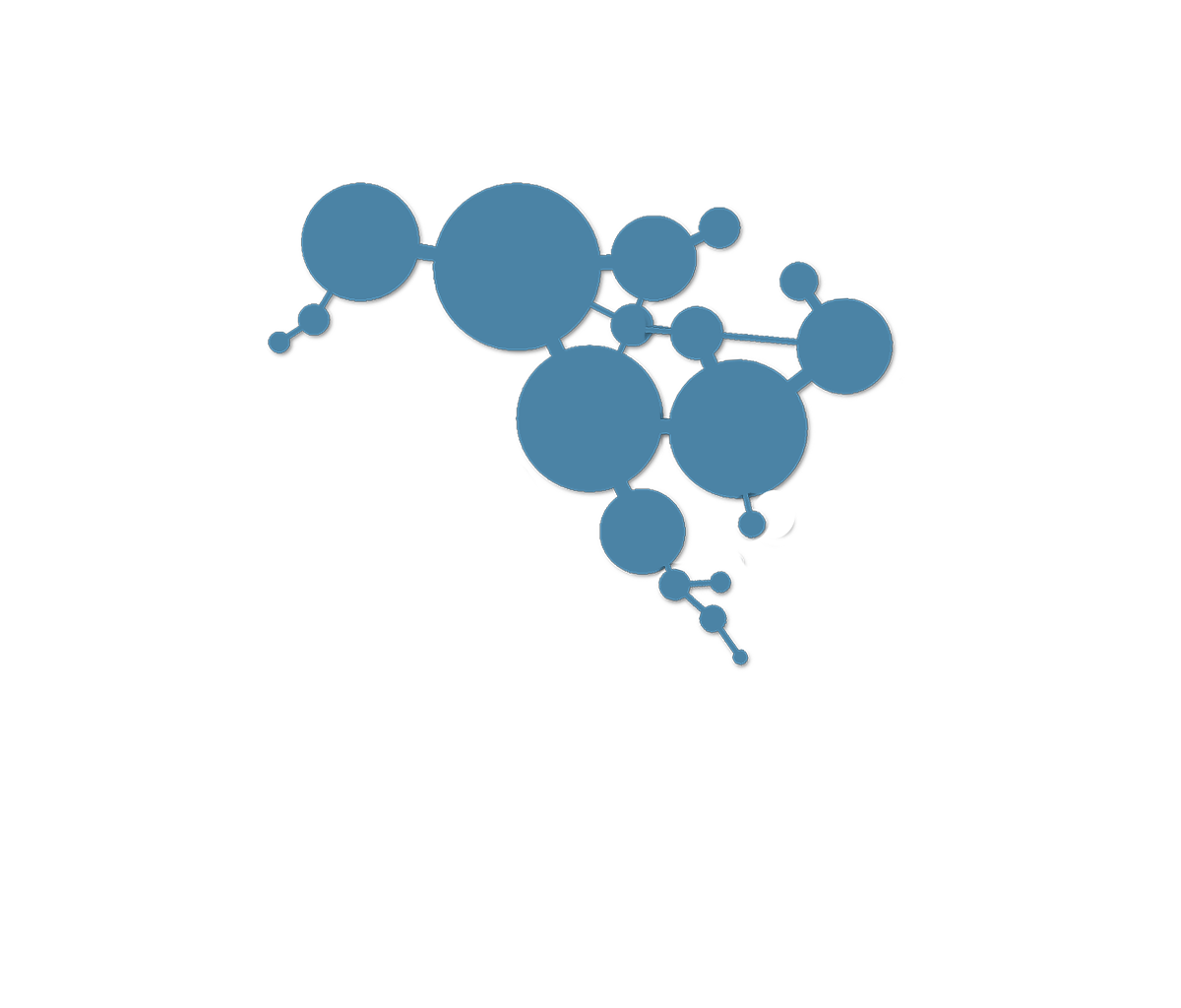Metabolomic Data Analysis Using MetaboAnalyst
This is the fifth module in the 2016 Informatics and Statistics for Metabolomics workshop hosted by the Canadian Bioinformatics Workshops. This lecture is by David Wishart from the University of Alberta.
How it Begins by Kevin MacLeod is licensed under a Creative Commons Attribution license (https://creativecommons.org/licenses/...)
Source: http://incompetech.com/music/royalty-...
Artist: http://incompetech.com/
Table of Contents:
00:10 -
00:56 - Learning Objectives
01:15 - A Typical Metabolomics Experiment
03:16 - 2 Routes to Metabolomics
03:31 - Metabolomics Data Workflow
05:11 - Data Integrity/Quality
06:46 - Data/Spectral Alignment
07:28 - Binning (3000 pts to 14 bins)
07:53 - Data Normalization/Scaling
09:40 - Data Normalization/Scaling
10:20 - Data QC, Outlier Removal & Data Reduction
12:08 - MetaboAnalyst
13:25 - MetaboAnalyst History
14:37 - MetaboAnalyst Overview
16:00 - MetaboAnalyst Modules
17:00 - MetaboAnalyst Modules
18:03 - Example Datasets
19:09 - Example Datasets
19:34 - Metabolomic Data Processing
19:44 - Example Datasets
19:45 - Example Datasets
19:45 - MetaboAnalyst Modules
19:48 - Example Datasets
19:48 - Example Datasets
19:49 - Metabolomic Data Processing
19:51 - Common Tasks
20:38 - Select a Module (Statistical Analysis)
20:48 - Common Tasks
20:49 - Metabolomic Data Processing
20:49 - Example Datasets
20:49 - Example Datasets
20:55 - Example Datasets
20:55 - Metabolomic Data Processing
20:56 - Common Tasks
20:56 - Select a Module (Statistical Analysis)
21:07 - Data Upload
21:37 - Alternatively …
22:18 - Data Set Selected
23:24 - Data Integrity Check
24:11 - Data Normalization
25:51 - Data Normalization
26:28 - Data Normalization
27:40 - Normalization Result
29:49 - Data Normalization
29:59 - Normalization Result
30:03 - Data Normalization
30:14 - Normalization Result
30:15 - Data Normalization
30:15 - Data Normalization
30:16 - Data Normalization
30:21 - Data Normalization
30:21 - Data Normalization
30:36 - Normalization Result
31:24 - Data Normalization
31:26 - Next Steps
31:37 - Quality Control
32:41 - Visual Inspection
33:36 - Outlier Removal (Data Editor)
34:01 - Noise Reduction (Data Filtering)
35:09 - Noise Reduction (cont.)
35:36 - Data Reduction and Statistical Analysis
36:07 - Common Tasks
36:30 -
37:00 - ANOVA
37:31 - ANOVA
38:58 - View Individual Compounds
39:46 - What’s Next?
40:06 - Overall Correlation Pattern
41:20 - High Resolution Image
41:53 - What’s Next?
42:40 - Pattern Matching
43:29 - Pattern Matching (cont.)
44:57 -
45:16 - Pattern Matching (cont.)
45:17 - Pattern Matching
45:32 - Pattern Matching (cont.)
45:34 -
45:55 - Multivariate Analysis
46:58 - PCA Scores Plot
47:40 - PCA Loading Plot
47:58 - PCA Scores Plot
48:15 - PCA Loading Plot
49:14 - 3D Score Plot
50:28 -
51:06 - 3D Score Plot
51:07 - PCA Loading Plot
51:08 - PCA Scores Plot
51:46 - PCA Loading Plot
51:47 - 3D Score Plot
51:49 -
52:30 - Multivariate Analysis
52:54 - PLS-DA Score Plot
53:23 - Evaluation of PLS-DA Model
55:19 - Important Compounds
57:07 - Model Validation
58:07 -
58:16 - Hierarchical Clustering (Heat Maps)
58:34 - Heatmap Visualization
58:48 - Heatmap Visualization (cont.)
59:16 - What’s Next?
59:28 - Heatmap Visualization (cont.)
59:29 - Heatmap Visualization
59:32 - Heatmap Visualization (cont.)
01:00:18 - What’s Next?
01:02:45 - Download Results
01:02:56 - Analysis Report
01:03:21 - Select a Module (Enrichment Analysis)
01:03:29 - Metabolite Set Enrichment Analysis (MSEA)
01:04:18 - Enrichment Analysis
01:04:56 - MSEA
01:05:09 - The MSEA Approach
01:05:18 - Data Set Selected
01:06:02 - Start with a Compound List for ORA
01:06:10 - Upload Compound List
01:06:38 - Perform Compound Name Standardization
01:06:55 - Name Standardization (cont.)
01:07:11 - Select a Metabolite Set Library
01:07:46 - Result
01:08:39 - Result (cont.)
01:09:04 - The Matched Metabolite Set
01:09:18 - Phenylalanine and Tyrosine Metabolism in SMPDB
01:09:42 - Single Sample Profiling (SSP)
(Basically used by a physician to analyze a patient)
01:10:14 - Concentration Comparison
01:10:34 - Concentration Comparison (cont.)
01:11:13 - Quantitative Enrichment Analysis (QEA)
01:11:30 - Result
01:11:47 - The Matched Metabolite Set
01:12:02 - Select a Module (Pathway Analysis)
01:12:09 - Pathway Analysis
01:12:57 - Data Set Selected
01:13:03 - Pathway Analysis Module
01:13:09 - Data Upload
01:13:22 - Perform Data Normalization
01:13:38 - Select Pathway Libraries
01:13:54 - Perform Network Topology Analysis
01:14:04 - Pathway Position Matters
01:14:44 - Which Node is More Important?
01:14:54 - Pathway Position Matters
01:14:56 - Which Node is More Important?
01:15:01 - Pathway Visualization
01:17:15 - Pathway Visualization (cont.)
01:17:28 - Pathway Impact
01:17:43 - Result
01:17:54 - Select a Module (Biomarker Analysis)
01:18:58 - Biomarker Analysis
01:19:37 - Select Test Data Set 1
01:19:42 - Data Set Selected
01:20:34 - Perform Data Integrity Check